A tour of scientist gem
Schools with large amounts of families complain that when visiting the Families roster, the request is too slow to use and even times out. This is a tour about how we made a performance boost while keeping the functionality work as normal.
Bottleneck
As the problem is repeatable, the bottleneck is easily found by checking logs. It is worth mentioning that, Flame Graph is a better way for the job1.
The bottleneck is a method fetching ancestor ids of all families to support the “Select all families” functionality, which allows users to select families from all pages.
@families.map { |family| family.valid_ordered_ancestors.map(&:id).join(',') }
Notice valid_ordered_ancestors is an association, it means 1000 SQLs for 1000
families, causing the duration increasing linearly with the families count.
First thought
As the method returns ids only, after talking with my team leader, we decided to
add a column to cache the ids of valid_ordered_ancestors, named
cached_valid_ordered_ancestors_ids, and update it in the after_touch
callback:
serialize :cached_valid_ordered_ancestors_ids, Array
after_touch :update_cached_valid_ordered_ancestors_ids
def update_cached_valid_ordered_ancestors_ids
self.cached_valid_ordered_ancestors_ids = valid_ordered_ancestors.ids
self.save!
end
Then the solution comes out:
@families.map { |family| family.cached_valid_ordered_ancestors_ids.join(',') }
Great! It reduced SQLs from 1000 to 1. Problem resolved!
But there is a risk: if the column is not updated in all cases, the method will return the wrong result, which is never expected. Because of this, we want to try the solution on production before switching to it while breaking the original functionality.
Be a “scientist”
The gem scientist comes to rescue. Here is the description:
🔬 A Ruby library for carefully refactoring critical paths.
It provides the ability to run experiment code on production while keeping the original functionality.
As a scientist, one of the ways to improve an existing solution is to make an controlled experiment2:
- Set two groups for the experiment: existing solution as the control group (control), new solution as the experimental group (candidate);
- Use control group’s result as baseline, then compare the candidate’s result with it to see whether the new solution is expected and better.
This concept can be seen in the following code examples.
Solution 1
First, include the Scientist module, it provides DSLs for defining experiments:
include Scientist
science 'family-roster-collect-ids' do |e|
# control group, old way
e.use { @families.map { |family| family.valid_ordered_ancestors.map(&:id).join(',') } }
# candidate group, new way
e.try { @families.map { |family| family.cached_valid_ordered_ancestors_ids.join(',') } }
end
To enable the experiment, it also requires us to provide an implementation of
Scientist::Experiment. I put it in config/initializers/scientist.rb:
require "scientist/experiment"
class ScientistExperiment
include Scientist::Experiment
attr_accessor :name
def initialize(name)
@name = name
end
def enabled?
true
end
def raised(operation, error)
super
end
def publish(result)
puts "science.{name}" # science.family-roster-collect-ids
puts result.matched? # whether results equal in both ways
puts result.control.duration # old way duration
puts result.candidates.first.duration # new way duration
end
end
Two methods need to notice:
enabled?returns true to enable the experiment;publishexposes the experiment results so you can send them to console, files or error monitors.
We deploy the code to production to collect the results for comparison and see whether the new solution is usable.
The results for a school having 4000 families:

Another school having 8000 families:

This solution is 3-4x faster but still not enough.
Solution 2
I assume it can be further improved if we return the joined ids directly, so
another column named cached_valid_ordered_ancestors_ids_str is added to store
the joined string.
def update_cached_valid_ordered_ancestors_ids
cache_ids = valid_ordered_ancestors.ids.dup
self.cached_valid_ordered_ancestors_ids = cache_ids
self.cached_valid_ordered_ancestors_ids_str = cache_ids.join(',')
self.save!
end
To confirm the assumption, I made a benchmark with benchmark-ips:
require 'benchmark/ips'
Benchmark.ips do |x|
x.config(:time => 3, :warmup => 0)
@collection = Family.all
# solution 1
x.report("map") do
@collection.map { |f| f.cached_valid_ordered_ancestors_ids.join(',') }
end
# solution 2
x.report("collect") do
@collection.collect(&:cached_valid_ordered_ancestors_ids_str)
end
x.compare!
end
and executed it with 600 families and the result is:
Calculating -------------------------------------
map 980.811 (±21.7%) i/s - 2.618k in 2.997818s
collect 4.545k (±10.5%) i/s - 13.288k in 2.992205s
Comparison:
collect: 4545.4 i/s
map: 980.8 i/s - 4.63x (± 0.00) slower
collect is 4.63 times faster than the map, it is really exciting!
Then the solution is changed to the following and deployed to production:
include Scientist
science 'family-roster-collect-ids' do |e|
# control group, old way
e.use { @families.map { |family| family.valid_ordered_ancestors.map(&:id).join(',') } }
# candidate group, new way
e.try { @collection.collect(&:cached_valid_ordered_ancestors_ids_str) }
end
The results for the 8000 families school, from 30 seconds to 0.01, it’s a 3000x boost! 💪
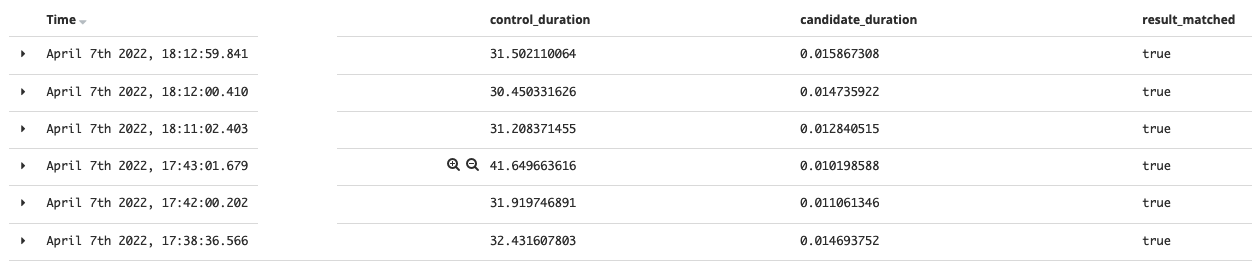
The school having 4000 families:

By using this solution, the method time comes to almost the same level regardless of the families count.
Observation
After running the experiment for two weeks, we can see performance improvement without result mismatch, so we switched to the new solution.
Conclusion
In this case we benefit much from scientist. It also has its limitation, for example, it may be not suitable if the results are not comparable, or you can not make decision just from the results. But when it is suitable, you can enjoy writing code with calm and confidence.